Impact of Different DNA Isolation Approaches on PCR Analysis of FFPE Tissue Samples
Laboratory of Molecular Pathology, Institute of Pathology, Centre Hospitalier Universitaire Vaudois and University of Lausanne, CH-1011 Lausanne, Switzerland
Publication Date: 2010
Abstract
In this study we compare methods for purification of DNA from FFPE tissue. DNA isolated using the Maxwell® 16 Instrument and the Maxwell® 16 Method performed better in downstream PCR when compared to traditional phenol:chloroform or a commercial spin-column extraction method.
Introduction
Isolation of genomic DNA from archival formalin-fixed and paraffin-embedded (FFPE) tissues is a critical step for studies involving pathological samples (1) (2) . The challenge is obtaining the maximum amount of DNA from a minimum of FFPE tissues with a quality level sufficient to perform all the necessary analyses. Other parameters such as time of the procedure, automation and cost of the DNA extraction also must be considered. A new technique of extraction with magnetic particles (3) using the Maxwell® 16 Instrument was tested and compared to two other procedures: a classical liquid-liquid extraction technique with phase separation (phenol:chloroform extraction), and a spin column-based nucleic acid purification (Qiagen extraction).
"With FFPE samples, PCR amplification is highly dependent on the size distribution and the quality of the extracted DNA."
Methods
DNA extractions were performed from consecutive 7µm FFPE tissue sections. A deparaffinization followed by coloration with toluidine blue allowed microdissection before the extraction to separate the tumor from the normal tissue. Ten cases were analyzed. After the microdissection step, the number of samples was multiplied by two: one tube for the normal tissue and another one for the tumor.
Isolation of genomic DNA from consecutive tissue sections was performed with three different protocols. We used the classical phenol extraction with a homebrew protocol, followed by ethanol precipitation. The Qiagen method was performed following the instructions of the DNeasy® Blood & Tissue Kit (Qiagen Cat.# 69504) procedure, which is largely used in diagnostic laboratories. Finally, an automated genomic DNA extraction using the Maxwell® 16 Instrument was performed. First, we used the Maxwell® 16 FFPE Tissue LEV DNA Purification Kit, but we observed that the chemistry had a limited DNA capture making it impossible to extract more than about 1μg of DNA. In order to extract more DNA from FFPE samples, the protocol of the Maxwell® 16 Tissue DNA Purification Kit (Cat.# AS1030) was adapted. The microdissected material was digested overnight at 70°C with 180μl of Incubation Buffer and 20μl of Proteinase K. After addition of 400µl of lysis buffer (Cat.# A8261), the solution was transferred into well #1 of the cartridge, and the procedure was followed as indicated in the protocol (TM284).
NanoDrop® quantification and p53 quantitative PCR (qPCR) were used as controls for quantity and quality of each DNA extracted. Moreover, 1% agarose gels were performed to estimate size distribution of extracted DNA.
Results
Comparison of Total DNA Quantity Collected
The total amount of DNA extracted with the different approaches from 10 of the 20 samples is shown in Figure 1. The highest quantities of genetic material were obtained with the Qiagen kit, followed by the phenol extraction procedure. Yield using the Maxwell® method was, on average, about 1.8 times lower than the phenol extraction method and about 2 times lower than the Qiagen extraction method. Automated extraction using the Maxwell® system was 1 to 4 times less efficient than the Qiagen extraction method and 1 to 3 times less efficient than the phenol extraction method. In some cases, the three different protocols gave similar yields, as in sample 1. In others, the yields were dramatically different, as in sample 10.
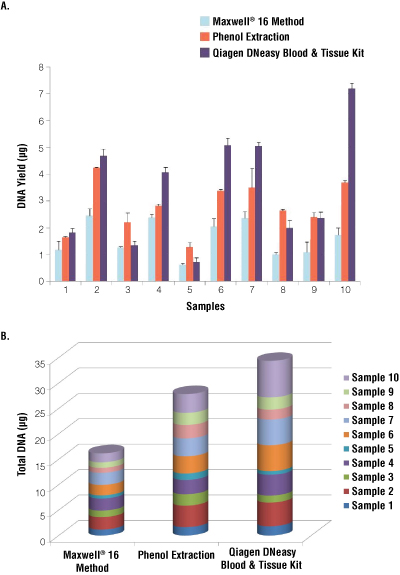 Figure 1. Comparison of 3 different methods of DNA extraction by comparison of the quantity of nucleic acids (ng).
Figure 1. Comparison of 3 different methods of DNA extraction by comparison of the quantity of nucleic acids (ng).
Panel A. Quantity of DNA collected for each sample (µg). Panel B. Cumulative DNA yield from ten samples. NanoDrop® quantification was performed with 2µl of DNA.
qPCR Amplification Efficiency
With FFPE samples, PCR amplification is highly dependent on the size distribution and quality of the extracted DNA, which is why amplifications using the same amount of DNA can produce different results. The quality of the DNA was tested by amplifying a 202bp fragment of the p53 gene using quantitative PCR. qPCR amplifications were performed with the same relative volume of eluate (1/50 of the final extraction volume). The relative amplification of the samples was calculated by comparison to a DNA control. PCR amplifications from DNA extracted using the Maxwell® protocol were 1 to 9 times more efficient than PCR amplifications using DNA from phenol or Qiagen extraction (Figure 2, Panel A). PCR from DNA extracted with the Maxwell® System can be as much as three times more efficient than PCR of DNA purified using phenol extraction and 2 to 9 times more efficient than DNA purified using the Qiagen extraction method.
This means that even if the DNA yield from a Maxwell® System extraction is lower than that of the two other techniques, PCR using the Maxwell®-extracted DNA is highly efficient, and therefore the results reflect the superior purity of the DNA extracted with the Maxwel® 16 method.
When the results are normalized by input amount of DNA, purification using the Maxwell® System was even more efficient (Figure 2, Panel B). Indeed, the qPCR of DNA template purified using the Maxwell® System was 1 to 10 times more efficient than qPCR of DNA template purified using the phenol extraction method. The difference in qPCR efficiency is even more noticeable when comparing DNA purified using the Qiagen system with qPCR of DNA template purified using the Maxwell® System being 5 to 24 times more efficient. In summary the DNA extracted using the Maxwell® System was 4 or 13 times more efficient, on average, than DNA extracted using the phenol or Qiagen protocols, respectively.
Additionally, we performed agarose gel electrophoresis to determine size distribution of isolated DNA data not shown). Phenol extraction recovered more DNA fragments smaller than 200bp compared to DNA purification using the Maxwell® System. This could explain why the PCR was less efficient with phenol-extracted DNA. The size distribution of DNA purified by the Qiagen and Maxwell® procedures was similar and therefore does not explain the change in the quality of the extracted DNAs.
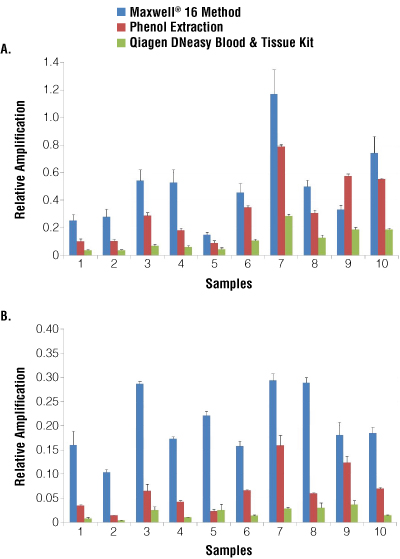 Figure 2. Quality of isolated genomic DNA determined by qPCR analysis.
Figure 2. Quality of isolated genomic DNA determined by qPCR analysis.
Panel A. Comparison of the relative PCR amplification for each method with the same initial volume in the PCR. Panel B. Comparison of the relative PCR amplification for each method with the same amount of extracted DNA in the PCR (10ng of genomic DNA per reaction). The PCR amplifications were performed in duplicate on Rotor-Gene® 6000 with 500nM of primers, 2mM MgCl2, 250μM dNTP, 5μM Syto®9, 0.5 units of Platinum Taq® Polymerase (Invitrogen). Relative amplification was calculated by normalizing the amplification of DNA samples to that of a control sample.
Assessment of Other Parameters
For clinical applications, the risk of contamination occurring from environmental sources or cross-contamination between the samples must to be reduced to an absolute minimum. The Maxwell® System is designed for automated isolation of genomic DNA. The extraction procedure requires minimal manipulation and thus, minimizes the risk of contamination. On the contrary, the risk is relatively high with the phenol extraction protocol due to repetitive changes of tubes during aqueous phase recoveries. For Qiagen extraction, the risk of contamination is slightly higher than with Maxwell® because there are more pipetting steps, and there is a risk of solutions splashing out of the columns.
The rapidity of the procedure is another important parameter to take into account. The phenol extraction, which comprises an overnight ethanol precipitation, is the longest procedure. Even though the lengths of the protocols are similar, the automated Maxwell® System method requires less hands-on time than the Qiagen extraction.
The last parameter to evaluate was the cost of DNA isolation with the three different protocols. The cheapest method is undoubtedly the phenol extraction. Qiagen and Maxwell® purifications are about 3 and 3.5 times more expensive, respectively.
Considering the following parameters—the quantity and the quality of the isolated DNA, the rapidity and the cost of the extraction procedure and the risk of cross-contamination—the Maxwell® protocol gets the most points based upon these five criteria (Figure 3). The Maxwell® System extraction is the method of choice for quality of isolated DNA, rapidity of the procedure and lowest risk of cross-contamination.
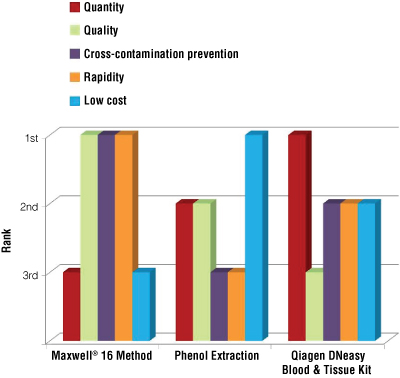 Figure 3. Comparison of different methods of extraction depending on 5 criteria: DNA quantity, DNA quality, ability to prevent cross-contamination, rapidity of the extraction procedure and cost for DNA isolation.
Figure 3. Comparison of different methods of extraction depending on 5 criteria: DNA quantity, DNA quality, ability to prevent cross-contamination, rapidity of the extraction procedure and cost for DNA isolation.
In this representation, for each criterion, the first rank was allocated to the extraction method.
Summary
The automated DNA extraction with the Maxwell® 16 Instrument is the method of choice for isolating genomic DNA from FFPE samples. Despite the fact that the amount of isolated DNA is the lowest of the three methods compared here, the amplification of the extracted DNA is much more efficient. This means that less DNA is needed to perform PCR. The isolated DNA can be diluted to a ratio of 1/5 to 1/20, depending on the samples, and therefore, 5 to 20 more PCR amplifications can be performed per sample, which is important for analysis of limited biological samples.
We hypothesize that the principle of DNA extraction with magnetic beads, as performed with the Maxwell® 16 System, makes it possible to isolate DNA of higher quality than those obtained with other protocols. The purity of the DNA is higher probably because of reduced contamination with RNA, proteins and PCR inhibitors.
Related Protocols
Article References
- Okello, J.B. et al. (2010) Comparison of methods in the recovery of nucleic acids from archival formalin-fixed paraffin-embedded autopsy tissues. Anal. Biochem. 400, 110–7.
- Bonin, S. et al. (2010) Multicentre validation study of nucleic acids extraction from FFPE tissues. Virchows Arch. 457, 309–17.
- Berensmeier, S. (2006) Magnetic particles for the separation and purification of nucleic acids. Appl. Microbiol. Biotechnol. 73, 495–504.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Bougel S. and Benhattar J. Impact of Different DNA Isolation Approaches on PCR Analysis of FFPE Tissue Samples. [Internet] 2010. [cited: year, month, date]. Available from: https://www.promega.com/resources/pubhub/impact-of-different-dna-isolation-approaches-on-pcr-analysis-of-ffpe-tissue-samples/
American Medical Association, Manual of Style, 10th edition, 2007
Bougel S. and Benhattar J. Impact of Different DNA Isolation Approaches on PCR Analysis of FFPE Tissue Samples. Promega Corporation Web site. https://www.promega.com/resources/pubhub/impact-of-different-dna-isolation-approaches-on-pcr-analysis-of-ffpe-tissue-samples/ Updated 2010. Accessed Month Day, Year.
Maxwell is a registered trademark of Promega Corporation. DNeasy is registered trademark of Qiagen GmbH Corporation. NanoDrop is a registered trademark of Thermo Fisher Scientific. Platinum is a registered trademark of Life Technologies. Rotor-Gene is registered trademark of Corbett Research. Syto is a registered trademark of Life Technologies.
 Meet Maxwell®, a fast, reliable way to purify DNA.
Meet Maxwell®, a fast, reliable way to purify DNA.